TBMT42 Lab 2: parameter estimation and model uncertainty analysis¶
This is the second computer exercise in the course TMBT42 Systems biology, Digital Twins and AI. In this computer exercise we will look at how we can combine mathematical models with experimental data to gain more insight in to the biological system. This computer exercise in developed for MATLAB however, you are free to implement it in any environment of your choosing. Although, please note that the practical instructions below will focus on implementation in MATLAB.
Throughout the lab you will encounter different "admonitions", which as a type of text box. A short legend of the different kinds of admonitions used can be found in the box below:
Different admonitions used (click me)
Background and Introduction
Useful information
Guiding for implementation
Here you are tasked to do something
Reflective questions
To start with here are some instructions for what you'll need to install before starting with the lab. Note that if you have done the previous modules in the TMBT42 course you will likely already meet many of these requirements.
Before you start with this computer exercise, please ensure that you have the following :
- A suitable environment (MATLAB, Python, R etc.) installed and running on the computer you will be working on. For installation and setup of MATLAB, please see Matlab installation and Matlab packages for how to install additional matlab packages.
-
Suitable toolbox packages installed. You will need tools for optimisation algorithms, statistical testing, and random sampling. In MATLAB you'll want the following toolboxes installed:
- "Global optimization Toolbox"
- "Statistics and Machine Learning Toolbox"
-
A suitable setup to simulate ODE-models installed. In MATLAB you should use IQMtools toolbox.
- If you are using IQMtools for MATLAB you will need a C-compiler installed in Matlab
For this module you will receive a few pre-made scripts for Matlab. You can download these files here Lab2 files. Download and extract the files to a suitable locations.
As stated above, you are free to use any software environment however, you will need to redo the pre-made scripts and find compatible tools yourself.
Introduction¶
Read the introduction "hidden" below.
Practical Information (Click to expand)
Purpose of the lab¶
This computer exercise aims to introduce some of the techniques used to analyse if a mathematical model can accurately describe an experimental system. To achieve this we will look at how to qualitatively adn quantitatively evaluate how well a model describes experimental data. We will investigate how statistical hypothesis testing can be used to determine if a model should be rejected or not. We will formulate and implement the optimisation problem that arises when we want to use experimental data to estimate our model parameters. Lastly, we will look at different techniques that can be used to evaluate the models uncertainty with regards to experimental data. In other words, to what degree can we trust different aspects of the model given som e specific data.
How to pass¶
At the very bottom of this page you will find a collapsible box with questions. To pass this lab you provide satisfying answers to the questions in this final box in your report.
Throughout this lab you encounter green boxes marked with the word "Task", these boxes contain the main tasks that should be performed to complete the lab. There are nine tasks in total, and completing these tasks should help you answer the questions at the end. The questions can be answered in a point-by-point format but each answer should be comprehensive and adequate motivations and explications should be provided when necessary. Please include figures in your answers where it is applicable.
Background
In this computer exercise we will work with a very simple model that describes how blood pressure changes with age. Hypertension or high blood pressure is a condition defined as having a systolic blood pressure (SBP) above 140 mmHg and diastolic blood pressure (DBP) above 90 mmHg. Individuals suffering from hypertension run an elevated risk of developing further complications such as heart failure, aneurysms, renal failure, and cognitive decline. In addition, it is commonly observed that blood pressure increases with age, thus understanding what this relationship loos like is crucial for comprehending the associated risks and identifying factors that influence blood pressure.
When investigating blood pressure one has to keep in mind that the blood pressure depends on the cardiac cycle which is a dynamic process governed by the heart. This means that for each heart beat there will be an initial high pressure phase, when the heart contracts, followed by a low pressure phase when the heart is refilled. The systolic blood pressure (SBP), is a measure of the pressure exerted on arterial walls during the contraction phase of the heart. The diastolic blood pressure (DBP) reflects the pressure in the arteries when the heart is at rest between beats. Lastly, the mean arterial pressure (MAP) is a calculated value that provides an estimate of the average pressure exerted on the arterial walls throughout the cardiac cycle. The MAP takes into account both the SBP and DBP, incorporating the time spent in each phase of the cardiac cycle. MAP is subsequently an important parameter because it reflects the perfusion pressure necessary to deliver oxygen and nutrients to organs and tissues. It is particularly relevant in assessing overall cardiovascular health and evaluating the adequacy of blood flow to various organs.
The boxes below describes the model and the data that we will be using to analyse these processes.
Model
The model used in this exercise is very simple and consists of two ordinary differential equations (ODEs) with a total of 6 parameters. The two states described by these ODEs are the SBP and DPB respectively.
The parameters \(k_{1,SBP}\) and \(k_{1,DBP}\) represents a combination of factors that are not significantly altered by the ageing process. These are typically inherent factors such as vascular tone or cardiac function.
The parameters \(k_{2,SBP}\) and \(k_{2,DBP}\) represents a combination of factors that are directly connected to the ageing. These could be factors such as increased arterial stiffness, reduced arterial compliance, and altered vascular function, all of which can contribute to the observed age-related change in blood pressure.
The parameters \(b_{SBP}\) and \(b_{DBP}\) represents the influence of factors that contribute to a person's baseline blood pressure level. These factors may include genetic predispositions, lifestyle choices, environmental influences, and chronic health conditions that impact blood pressure regulation.
\(SBP_0\) and \(DBP_0\) are the initial values for the two states respectively.
Lastly, the MAP is calculated as the difference between the SBP and the DBP, divided by 3 and added to the DBP.
Experimental data
For this exercise we will consider data for the SBP, the DBP and the MAP measured at different ages in a fictional population. Figure 1 below provides an illustration of this data, and the relationship between age and blood pressure. The blue data points represents the SBP, the orange data points corresponds to the DBP, and the green data points represents the MAP. The data shows th that on average the SBP increases some linearly over the age span of 30 to 80 years, while the diastolic blood pressure increases at first but around age 50 it starts to decline. The MAP as expected represents the combined behaviour of both the SBP and DBP.
Figure 1: Figure 1 illustrates the experimental data for how the blood pressure increases with age for systolic blood pressure (SBP) in blue, diastolic blood pressure (DBP) in orange, and the mean arterial pressure (MAP) in green. The data is displayed as mean value with error bars indicating the standard error of the mean (SEM).
Parameter Estimation¶
Background: What is parameter estimation?
Parameter estimation is the process of estimating what model parameter values create the behaviour described by the experimental data. To evaluate whether our model can describe the observed data we need to find the parameter values that creates a good agreement between the data and the model simulation. If the agreement between model and data is not good enough assuming we have the best possible parameters, the hypothesis must be rejected. While it is possible to find this best combination of parameters by manually tuning the parameters. This is an extremely labour intensive process and would almost certainly take way too much time. Especially for more complex models where non-linear dynamics are introduced between the model parameters and the model behaviour. Instead, we can find this best agreement using optimisation methods.
An optimisation problem generally consists of two parts, an objective function with which a current solution is evaluated and some constraints that define what is considered an acceptable solution. We will look at how we can formulate the objective function in Task 2 below. As for our constraints these depend a bit on what type of model we are using but for the most part it will suffice with a set of lower and upper bounds (lb/ub) on the parameter values as constraints. The optimisation problem can then typically be formulated as something a kin to the equations below.
Where we are minimizing the value of the objective function \(v\) which will depend on the parameter values \(\theta\). With the constraints that \(\theta\) cannot be smaller than \(lb\), nor larger that \(ub\).
As the value of the objective function can scale non-linearly with the parameter values \(\theta\) the optimisation problem can be hard to solve, and the solution we find is therefore not guaranteed to be the best possible solution. We can for example find a solution which is minimal in a limited surrounding, a so-called local minimum, instead of the best possible solution – the global minimum. This relationship is visualised in Figure 2 where different sets of parameter values \(\theta\) yield different values of the objective function to form a landscape with various hills and valleys. There are methods to try to reach the global minimum, for example you can run the optimisation several times from different starting points and look for the lowest solution among all runs. Please note that there might be multiple solutions with equal values.
Furthermore, there exists many methods of solving an optimisation problem. If a method is implemented as an algorithm, it is typically referred to as a solver. Different solvers will use different techniques to find their way "downhill" in the landscape of the objective function, all attempting to reach the global minimum.
Figure 2: An illustration of how the objective-function value varies with different parameter values \(\theta\). The figure also highlights the difference between a local and a global minimum and how the different solutions correspond to a model simulation.
We will start by getting to know the system we will be working with. Firstly we will take a look at the simulating the blood pressure model and looking at our data. Then we will go into more detail of how we can conduct the parameter estimation analysis. The boxes below contain information to help you in this process with the specific tasks to complete indicated by the green boxes, while blue boxes contain more general information about the different concepts we'll encounter.
Simulate the blood pressure model
In this exercise you will not need to formulate and implement the model yourself. Since you should already have gone through these steps in the previous computer lab we will skip them here. Instead, in the pre-made files you have downloaded, you'll find a model text file with the model equations already implemented. You will also find a matlab function called "SimulateBloodPressure.m", this function simulates the model.
NOTE That you will need to run the mex-compilation on your own computer before running the model simulation! since we do not plan to make any changes to the model file, you should only need to do this once. Here is an example of how to compile the model mex-file.
modelName = 'bp_age';
model = IQMmodel([modelName '.txt']); % Create the iqm model object
IQMmakeMEXmodel(model)% compile the mex files
To call the provided model simulation function you'll need the model name as a string ('bp_age'), a parameter vector consisting of 8 scalar values (the 6 parameters and the initial value for each state), and a vector of time points for which to simulate the blood pressure. For these inputs the function call will return a simulation structure as can be seen in the example below where
modelName = 'bp_age';
time = [1:10];
modelSimulation = SimulateBloodPressure(modelName, parameterValues, time)
modelSimulation =
struct with fields:
time: [1 2 3 4 5 6 7 8 9 10]
states: {'SBP' 'DBP'}
statevalues: [10×2 double]
variables: {'MAP' 'age' 'SBP_vs_IC' 'DBP_vs_IC'}
variablevalues: [10×4 double]
reactions: []
reactionvalues: []
Should the simulation fail for what ever reason the function will return a similar structure but only consisting of the fields "statevalues" and "variablevalues". Additionally, all values in these matrices will be 1e20.
modelSimulation =
struct with fields:
statevalues: [10×2 double]
variablevalues: [10×4 double]
SBP and DBP you can call the field statevalues, and to access the simulated values for the variable MAP you would call variablevalues. Note that these fields contain matrices with rows equal to the number of time points used for the simulation, here the 10 rows correspond to the time points time = [1:10]. The columns of these matrices correspond to states and variables respectively i.e. the first column in statevalues contains the simulated values for SBP, the second column in statevalues contains the simulated values for DBP, and the first column in variablevalues contains the simulated values for MAP.
For example:
SBP = modelSimulation.statevalues(:,1);
Task 1: Plot a model simulation of the state SBP along with data for SBP.
To start out you should make a figure where you plot the data for SBP and a simulation for SBP in the same plot.
- The data should be plotted as error bars indicating the mean and SEM for each data point.
- It should be clear what is the data and what is the simulation.
- The figure should have clear legends, labels and a title.
Use the following parameter values for the initial simulation.
| Parameter values |
|---|
| k1_SBP = 1 |
| k2_SBP = 0 |
| k1_DBP = 1 |
| k2_DBP = 0 |
| bSBP = 100 |
| bDBP = 50 |
| IC_SBP = 115 |
| IC_DBP = 60 |
Some useful MATLAB commands to help you plot are:
`errorbar`, `xlabel`, `ylabel`, `title`, `plot`, `hold on`
You are free to use any plot function you may have designed in the previous lab to accomplish this task. If you do not have a suitable plot function it is highly recommended that you build one as plotting data and simulation will be a recurring element of this lab.
Save you figure in a suitable location with a useful name. A useful name would be something short and concise that still informs a user what the figure might contain, e.g. Data_and_initialSimulation.png.
Now we want to evaluate how well the agreement between our model simulation and the data really is. We can already do this qualitatively by inspecting the figure you've just made. How good is the agreement? However, in order to use the computational tools we have available we need a way to describe this agreement between model and data quantitatively, i.e. we would like t assign a number to how good the agreement is.
Quantitative Agreement to data
To better evaluate the model we want to test it quantitatively. Let us first establish a measure of how well the simulation can explain data, also known as the cost of the model. We use the following formulation of this cost:
Where \(v(\theta)\) is the cost, given a set of parameters \(\theta\). \(y_t\) is the mean values of the data points and \(SEM_t\) is the standard error of the mean at each respective time point \(t\), and \({\hat{y}}_t\) is model simulation at time \(t\). Here, \(y_t-\hat{y}_t\left(\theta\right)\), is commonly referred to as residuals (Fig 3), and represent the difference between the simulation and the data. In the equation above, the residuals weighted with the SEM values, squared and summed over all time points \(t\). The residuals gives us the quantitative difference between the mean value of the data \(y_t\) and the simulated value \(\hat{y}_t\left(\theta\right)\) of each time point. Dividing the residuals by with the standard deviation \(\sigma\) , or SEM if mean values are used, ensures that data points that are well determined contributes more to the total cost of the fit. In other words, data points with small values of \(\sigma\)/SEM will yield a larger quotient and thus be weighted more when evaluating the quantitative model agreement ot the data. Lastly, squaring the weighted residuals ensures that all residuals has a positive contribution to the total cost. Since some residuals will be positive \(y_t \gt \hat{y}_t\left(\theta\right)\) and some will be negative \(y_t \lt \hat{y}_t\left(\theta\right)\) we don't want these different sings to cancel each other out. Thus by squaring the values all residuals will have a positive contribution to the total cost.
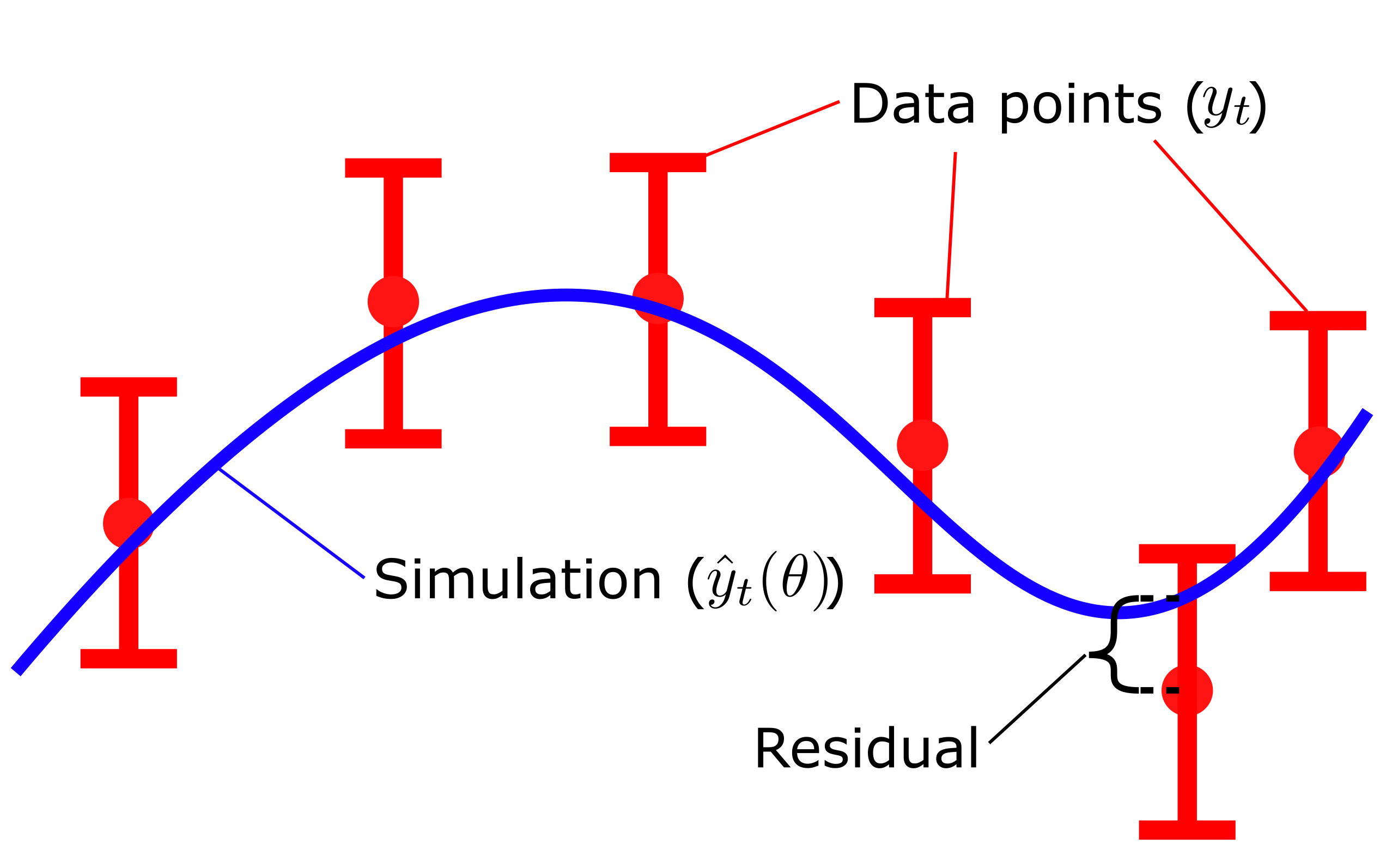 Figure 3: Residuals are the difference between model simulation and the mean of the measured data
Figure 3: Residuals are the difference between model simulation and the mean of the measured data
Task 2: Build a function that calculates the cost.
Build a function that takes the necessary inputs, simulates the model and calculates the agreement to data for that simulation.
Your cost function should:
- Take the parameter vector as the first input argument. This is to ensure compatibility at later stages.
- Run a model simulation for those specific parameter values. The simulation function will need the model name and a time vector in addition to the parameter values.
- Calculate the cost of the model simulation with respect to the data for SBP.
- Return the calculated cost as the only function output.
You should aim for the the following function structure between the cost function and simulation function.
sequenceDiagram
Main->>CostFunction: parameterValues, modelName, Data
CostFunction ->> SimulateBloodPressure: parameterValues, modelName, time
SimulateBloodPressure ->> CostFunction: modelSimulation
CostFunction ->> Main: costNote: MATLAB interprets operations such as *, /, ^ between vectors as vector operations e.g. a*b if a and b are vectors will result in vector multiplication. You likely want element wise operations. This can be solved by introducing a dot . before any such operation (such as changing / to ./) e.g. a./b will divide each element of a with the corresponding elements in b. Please note that this is not an issue for +, -, if the vectors are of the same size.
You might have to transpose modelSimulation when calculating the cost (but probably not). This can be done by adding a .' after the variable (e.g. modelSimulation.').
Now that you can quantitatively evaluate how well a given set of parameter values can describe the data, we can start to test different sets of parameters to see if we can the best possible agreement between model and data. As stated in the background we could do this for hand, testing different parameter values, but that seams strenuous. Rather, we can use the computer to do this for us. All we need is to find a good algorithm for how to test different parameter values.
Different optimisation solvers
As stated in the background section above, different solvers will use different techniques to find their way "downhill" in the landscape of the objective function, all attempting to reach the global minimum. In general the solvers will solve the the problem iteratively i.e. the solver will evaluate the objective function for the initial parameter values, permute the parameter values in a way that should lower the objective function value, and once again evaluate the objective function for the new parameter values. The way in which a solver permutes the parameter values is determined by the solvers optimisation algorithm and is not something we will go into too much detail here. One of the main challenges for these algorithms is to know if searching "uphill" might eventually result in a better solution and thus avoiding getting stuck in local minima. Some algorithms circumnavigates this challenge by only searching "downhill", thus never searching in a direction that yield a worse objective function value, these algorithms are called local optimisation algorithms. These local algorithms are typically faster but run a higher risk of getting trapped in a local minimum, unable to search any further. The solutions from these local algorithms are therefore somewhat dependent on the starting point i.e. the initial parameter values. Algorithms that do allow some degrees of searching "uphill" are called global optimisation algorithms and are generally a bit more computationally demanding but are also less sensitive to the starting point.
There are several different optimisation solvers built in to MATLAB and we will look at a few of them.
-
fmincon: This is a local solver for constrained non-linear optimisation problems. The function
fminconworks by minimizing an objective function where the parameters are subject to any combination of bound, linear, and non-linear constraints. The function can use various algorithm options such as interior-point, trust-region-reflective (requires a user-supplied gradient), active-set, and SQP (sequential quadratic programming). These algorithms are iterative and take steps towards a solution, adjusting the steps as necessary to improve the optimisation. -
particleswarm: This is an optimisation solver based on the particle swarm optimisation (PSO) algorithm. PSO is a computational method that optimises a problem by iteratively trying to improve a candidate solution with regard to objective function, inspired by the social behaviour of bird flocking or fish schooling. In
particleswarm, each potential solution, or 'particle', is moved around in the search-space according to simple mathematical formula over the particle's position and velocity, with a goal to find the best global solution. -
simulannealbnd: This solver is based on the so called simulated annealing algorithm. Simulated annealing is a probabilistic technique for approximating the global optimum of a given objective function. The name 'simulated annealing' comes from annealing in metallurgy, a technique involving heating and controlled cooling of a material to increase the size of its crystals and reduce their defects. Similarly, the
simulannealbndfunction in MATLAB provides an algorithmic approach to finding a good (not necessarily perfect) solution to an optimisation problem, and it's particularly useful for problems with many local minima where gradient-based methods might get stuck.¨
Remember that you can always use doc or help in matlab to get more information regarding a function
Task 3 Run a parameter estimation where you fit the blood pressure model to the data for the SBP
Write a script where you do do a parameter estimation. You are free to use any or multiple of the algorithms described above, or any additional optimisation solver that you are aware of.
Your goal is to investigate find a set of parameters that can describe the data for the SBP. You will want to ues the cost function you designed in Task 2 as the objective function.
To complete this task you should investigate the following points:
- Write a script that runs a parameter estimation.
- Make a figure where you plot the data for SBP and the model simulation for the best set of parameter values. How does this compare to the original plot from Task 1?
- Remember to save your figure!
- Does the choice of optimisation algorithm affect the solution of the best parameter values? Why/Why not?
- Does the initial parameter values affect the solution of the best parameter values? Why/Why not?
- Is there a significant difference in computational time between different optimisation algorithms?
- You should save the best set of parameter values you find. You do not need to save all parameter estimation solutions but you should save the best one.
Implementing fmincon in MATLAB
To use the function fmincon in MATLAB we will need a total of nine input arguments. These are
objectiveFunction- The objective function that the algorithm will try to minimize. For this exercise the cost function will be our objective function.x0- A vector of initial parameter values.Aandb- These are matrices that specify any linear inequality constraints of the optimisation problem. They will not be used in this example and simply set to be empty vectors[].Aeqandbeq- These are matrices that specify any linear equality constraints of the optimisation problem. They will not be used in this example and simply set to be empty vectors[].lbandub- These are vectors that specify the lower and upper bounds of the parameter values respectively. These must be vectors of equal size to the parameter vector.options- This is a variable that contains the options for the optimisation algorithm and are generated by the functionoptimoptions.
For the options variable you'll want to set the 'Display' property to the value 'iter' such that the iterations of the optimisation algorithm are displayed to the matlab console.
options = optimoptions('fmincon','Display','iter');
fmincon are of the most interest for us in this exercise these are:
- The parameter vector that yielded the best solution for the objective function.
- The best objective function value that the algorithm could find.
The implementation should look something like this:
[bestParameterValues, bestFval] = fmincon(myObjectiveFunction,initialParameterValues,[],[],[],[],lb,ub,[],options);
[]) in the line above are the inputs A,b,Aeq,beq, and nonlcon. neither of which are used in this example hence these inputs are empty.
Implementing particleswarm in MATLAB
To use the function particleswarm in MATLAB we will need a total of nine input arguments. These are
objectiveFunction- The objective function that the algorithm will try to minimize. For this exercise the cost function will be our objective function.nVariables- A scalar containing the number of parameters.lbandub- These are vectors that specify the lower and upper bounds of the parameter values respectively. These must be vectors of equal size to the parameter vector.options- This is a variable that contains the options for the optimisation algorithm and are generated by the functionoptimoptions.
For the options variable you'll want to set the 'Display' property to the value 'iter' such that the iterations of the optimisation algorithm are displayed to the matlab console.
options = optimoptions('particleswarm','Display','iter');
The first two outputs form particleswarm are of the most interest for us, these are:
- The parameter vector that yielded the best solution for the objective function.
- The best objective function value that the algorithm could find.
The implementation should look something like this:
[bestParameterValues, bestFval] = particleswarm(myObjectiveFunction,nVariables,lb,ub,options);
Implementing simulannealbnd in MATLAB
To use the function simulannealbnd in MATLAB we will need a total of nine input arguments. These are
objectiveFunction- The objective function that the algorithm will try to minimize. For this exercise the cost function will be our objective function.x0- A vector of initial parameter values.lbandub- These are vectors that specify the lower and upper bounds of the parameter values respectively. These must be vectors of equal size to the parameter vector.options- This is a variable that contains the options for the optimisation algorithm and are generated by the functionoptimoptions.
For the options variable you'll want to set the 'Display' property to the value 'iter' such that the iterations of the optimisation algorithm are displayed to the matlab console.
options = optimoptions('simulannealbnd','Display','iter');
The first two outputs form simulannealbnd are of the most interest for us, these are:
- The parameter vector that yielded the best solution for the objective function.
- The best objective function value that the algorithm could find.
The implementation should look something like this:
[bestParameterValues, bestFval] = simulannealbnd(myObjectiveFunction,x0,lb,ub,options);
NOTE that all of these solvers will assume that the objective function only requires one input argument i.e. the parameter values. If your cost function requires multiple input argument such as the data structure or model name, you can use an anonymous function to send additional input arguments to the objective function.
Enable passing one input variable to the objective function.
If you are using any of the solvers above, you will need to construct an anonymous function that can take only the parameter values as input, and pass along additional input arguments to the cost function.
Most solvers can only call the cost function with one input variable (the parameter values). We must therefore update our script so that we only pass one input from the script that runs the parameter estimation to the cost function.
One way to bypass this limitation is to update the cost function to only accept one input and instead get the data and select model by either loading the data and model name in the cost function, or by getting them in as global variables. However, both of these methods have issues and will slow down your code. A third (and better) alternative is to use an anonymous function. You can read more about anonymous functions here. Essentially, we create a new function which takes one input, and then calls our cost function` with that input as well as extra inputs.
How to implement an anonymous function in the example for simulannealbnd (identical for the other solvers):
Replace:
[bestParameterValues, bestFval] = simulannealbnd(myObjectiveFunction,x0,lb,ub,options);
With:
proxyObjFunc = @(p) myObjectiveFunction(p, dataStructure, modelName)
[bestParameterValues, bestFval] = simulannealbnd(proxyObjFunc,x0,lb,ub,options);
Here, proxyObjFunc acts as an intermediary function, passing along more arguments to the real objective function. To the line where you define proxyObjFunc you should add the inputs for your cost function, the inputs dataStructure and modelName are just examples.
As you will note this problem will be solved fairly quickly and thus it might be difficult to asses and compare the computational time. To alleviate this you can use MATLABs built-in tic and toc functionality.
How to use tic/toc in MATLAB
MATLAB's tic and toc functions are used to measure elapsed time. They are similar to a stopwatch; tic starts timing, while toc stops timing and reports the elapsed time. Here's a brief example:
tic;
[bestParameterValues, bestFval] = particleswarm(myObjectiveFunction,nVariables,lb,ub,options);
elapsedTime = toc;
fprintf('The operation took %.2f seconds\n', elapsedTime);
elapsedTime and printed in the fprintf statement.
You can use the following values for the parameters bounds. Remember that lb and ub should be vectors.
| Parameters | Lower bound | Upper bound |
|---|---|---|
| k1_SBP | -5 | 5 |
| k2_SBP | -5 | 5 |
| k1_DBP | -5 | 5 |
| k2_DBP | -5 | 5 |
| bSBP | 50 | 200 |
| bDBP | 20 | 120 |
| IC_SBP | 70 | 200 |
| IC_DBP | 50 | 120 |
You have now hopefully found one or several solutions that appear to provide a good agreement between the model simulation and the data. But how do we know if the solution is good enough?
How to know if the model fit is good enough?
There are several different statistical tests that can be used to reject models based on how well the model can describe given data. One of the most common, and the one we will focus on in this lab is the \(\chi^2\)-test. A \(\chi^2\)-test is used to test the size of the residuals, i.e. the distance between model simulations and the mean of the measured data
For data with normally distributed noise and a known standard deviation \(\sigma\), the cost function, \(v(\theta)\), will follow a \(\chi^2\) distribution, \(v(\theta)\) can thus be used as test variable in the \(\chi^2\)-test. Here, we test whether \(v(\theta)\) is larger than a certain threshold. This threshold is determined by the inverse of the cumulative \(\chi^2\) distribution function. In order to calculate the threshold, we need to decide significance level and degrees of freedom. A significance level of 5% (p=0.05) means a 5% risk of rejecting the model hypothesis when it should not be rejected (i.e. a 5 % risk to reject the true model). The exact number of degrees of freedom for this distribution can difficult to determine but for now we can use the number of terms being summed in the cost (i.e. the number of data points).
If the calculated \(\chi^2\)-test variable (i.e. the cost) is larger than the threshold, we must reject the null hypothesis for the given set of parameters used to simulate the model. The null hypothesis for the \(\chi^2\)-test is that the residuals (Figure 3) are small i.e. the model is in good agreement with the data. Rejecting this null hypothesis we say that the residuals are not small enough for us to believe that this specific model realisation and the data realisation of the system originates from the same distribution. this means that we do not believe that the system we are describing with our current model could have generated the measured data i.e. we are describing the wrong system.
Task 4: Evaluate your best parameter solution with a \(\chi^2\)-test
To determine if you should reject the current blood pressure model you should evaluate the best solution you've acquired with a \(\chi^2\)-test. Once you are confident that you have found a suitably good solution from the parameter estimation above, you should calculate the \(\chi^2\)-threshold for a significance level of 5% and 11 degrees of freedom (11 = number of data points for SBP).
Evaluate whether or not you should reject the blood pressure model with respect to the \(\chi^2\)-test!
Calculate the \(\chi^2\)-threshold in MATLAB
In order to calculate the \(\chi^2\)-threshold you can use the chi2inv function. chi2inv returns the inverse cumulative distribution function of the chi-square distribution with a given number of degrees of freedom at a given \(\alpha\)-level.
DoF = 11;
threshold = chi2inv(0.95,DoF);
You should now have a verdict of whether or not the blood pressure model is adequate for describing how the SBP changes with age. But we should also consider the DBP since this is an equally important measure for evaluating blood pressure.
Task 5: Repeat Tasks 1-4 considering both SBP and DBP
You should now modify you current implementation to do a parameter estimation where you consider the data for both the SBP and DBP. Realistically, this should only mean fairly minor modifications to the function where you plot the model simulation and the data, and the cost function.
To complete this task you should:
- Modify the plot function to plot both data for SBP and data for DBP, as well as the model simulations for both SBP and DBP. All in the same figure.
- Modify the cost function such that it considers both the residuals for the SBP and the residuals for DBP.
- Run a parameter estimation where you fit the model to both SBP and DBP simultaneously.
- Remember to save the best parameter estimation solution you find!
- Use the \(\chi^2\)-test to evaluate if the model should be rejected or not when both data for SBP and DBP are used.
- Plot the final fit with both data sets and model simulations in the same figure.
- Remember to save your figures!
Estimating model and parameter uncertainty¶
Now we have used the data for our system, here how the blood pressure changes with age, to create an estimation of what the parameter values of our model could be. This means that if we believe the data, we should also believe that our estimated value for e.g. \(k_{2,SBP}\) constitutes a reasonable value for the combination of factors that are directly connected to the ageing of our population. But this solution is only one out of a multitude of possible solutions, all of which would pass the \(\chi^2\)-test. Since we can only make a meaningful conclusion regarding the model when we reject the model, we cannot claim that the solution we found are better than any other solution that also passes the \(\chi^2\)-test. Hence, what we really want is the distribution of all parameter values that result in an acceptable model behaviour (Figure 4). In this next section we will look at a few different approaches to estimates such distributions, and what can we learn about the system by analysing the distributions of acceptable model behaviours.
The Importance of parameter uncertainty
As we have seen, in the field of systems biology, mathematical models play a crucial role in interpreting complex biological phenomena. Our models often contain various parameters representing biological characteristics such as different factors that affect the blood pressure. However, due to experimental limitations, inherent biological variability, and other sources of uncertainty, these parameters can rarely be known with absolute precision. This is why studying parameter uncertainties is crucial in systems biology.
Studying parameter uncertainties allows us to quantify the reliability of our model predictions. It is important to acknowledging that our models are not absolute but rather range within certain confidence intervals. Furthermore, understanding how sensitive the models are to variations in parameters can help identify key drivers of biological behaviour. This can inform experimental design, by focusing efforts on measuring the parameters that matter most, and lead to more robust and reliable predictions.
Moreover, a deep understanding of parameter uncertainties can reveal which parts of the system are well-defined and which parts are less known. This knowledge can be utilized to improve experimental designs, enhance the model by incorporating more data or refining assumptions, and ultimately increase our understanding of the biological system at hand. In this way, the study of parameter uncertainties not only bolsters the validity of systems biology models, but also acts as a guide for future research efforts.
Figure 4: An illustration of how the model uncertainty correlates to the objective-function landscape and the \(\chi^2\)-test. All parameter solutions that yields an objective function value that is below the \(\chi^2\)-threshold (blue shaded area) correspond to a collection of acceptable model simulation (blue shaded simulation)
Identifiable and unidentifiable parameters¶
The terms "identifiable" and "unidentifiable" are sometimes used to describe whether it's possible to uniquely determine these parameters based on the available data.
An "identifiable" parameter is one for which a unique estimate can be found from the data. That is, there is narrow range of parameter values that the can describe the data. The parameter can be well determined given the experimental data. This doesn't mean that we've necessarily found the true value of the parameter (due to experimental noise, model assumptions etc.), but rather that no other parameter values can produce a better fit to the data.
On the other hand, an "unidentifiable" parameter is one for which a wide range of different values could equally well fit the data. In other words, we can't pinpoint a unique estimate for the parameter from the data because several different parameter values produce essentially the same model behaviour. Unidentifiable parameters can present challenges in model calibration, as they lead to uncertainty in model predictions.
Markov Chain Monte Carlo (MCMC) sampling¶
Background: MCMC sampling
Markov Chain Monte Carlo (MCMC) methods are a class of algorithms in computational statistics used for sampling from a specific probability distribution. Monte Carlo sampling is a statistical technique that works by generating a large number of random samples from a probability distribution, and then approximating numerical results based on the properties of these samples. It's essentially a way to make numerical estimates using randomness. MCMC sampling expands on these concepts by selecting samples such that the next sample is dependent on the existing samples, creating a so called Markov chain. This will allows the algorithm to hone in on the quantity that is being approximated from the distribution, this is especially useful for problems with a large number of variables.
As a very simplified example, Imagine we have a large jar filled with marbles of different colours and we want to figure out what percentage of the marbles are red, blue, green, yellow, etc.. In other words, we want to find the distribution of coloured marbles in the jar. Imagine also that the jar is to large to count the marbles one by one. The principles of MCMC sampling could be used to estimate this colour distribution. We can construct a simple algorithm for solving this task:
- You reach into the jar and select a marble at random. You record the colour and put the marble back in the jar.
- You, again at random, pick another marble from the jar. If it's the same colour as the previous one, we put it back and pick another one. If it's a different colour, we record its colour and put it back in the jar. This "choosing based on the last choice" is the "Markov Chain" part.
- We repeat step 2 a lot of times, thousands or millions of times. This is the "Monte Carlo" part, using repeated random sampling to obtain results.
- Finally, we look at the colours we recorded. The proportion of each colour in our records gives us an estimate of the proportion of each colour in the jar.
The more samples we draw (the more marbles we pull out), the closer our estimated distribution gets to the actual distribution.
Please note that this is a very simplified analogy. Real MCMC methods can get quite complex, particularly when it comes to determining the rules for when to accept or reject the next sample, but the general idea remains the same.
For our purposes, we can use MCMC sampling to estimate the distribution of our model parameters or a specific model behaviour. Let's say that we want to approximate the parameter distributions that allow our model to explain the data. We can then use a MCMC sampling algorithm to select different parameter sets. We can then use a similar cost function as in the previous section to evaluate if the selected parameters describe the data adequately well, and if so we record the selected parameter set and move on. Thus by gathering enough samples, we can get an approximation of the possible parameter values that allow the model to still explain the data.
Task 6: Run a MCMC sampling for the blood pressure model
You should now run a MCMC sampling for the blood pressure models capability to describe the SBP and DBP data. To complete this task you should:
- Implement a suitable MCMC sampling method of your choice.
- Run the MCMC sampling
- Make a figure that plot the parameter distributions for all solutions that passes the \(\chi^2\)-test with respect to both the SBP and DBP data
- Evaluate the parameter distributions in your figure.
- To the best of your understanding, are the parameter distributions reasonable? Why/why not?
- What can you determine from the parameter distributions? Do any of the parameter appear to be identifiable?
Saving acceptable parameter values
To start with we will look at how to record and save parameter samples that are good enough to pass the \(\chi^2\)-test. One way to do this is to save acceptable parameter values to a separate file.
FileName = 'acceptableParameters.dat'; % a suitable file name of your choice.
fileID = fopen(FileName,'wt'); % open the file so that matlab can write to it.
You also want to slightly modify your cost function such that it takes the file handle fileID as an additional input. You then want to in your cons function introduce an if clause that checks if the current parameter values yield a solution that is below the \(\chi^2\)-threshold. If this is true, you want to write those parameter values to the file.
Here is one example of how this can be done.
if and(~isempty(fileID),fval <= chi2inv(0.95,22)) % if the variable fileID is not empty and the variable fval is smaller than the chi2-threshold
fprintf(fileID,'%4.10f %10.10f ',[fval, parameterValues]); fprintf(fileID,'\n');
end
fileID is not empty and the variable fval is smaller than the \(\chi^2\)-threshold only if both of these conditions are true the if clause is triggered. The if clause then prints the fval and the parameterValues to the file specified by fileID. The line concludes by printing a "new line" format to the file so that additional entries are not printed on the same line.
Remember to close the file once you will not write any more to the file.
fclose(fileID)
Implementing a simple MCMC sampling method in matlab
Now that you can save the acceptable parameters that are encountered during the sampling. We will look at how to implement a simple MCMC sampling method. You will want to implement this analysis in a new matlab script, separate from you parameter estimation. The following code implements a very simple MCMC algorithm. You can copy this code for your own implementation. Please note that you will need to add some code that
- Load your data structure.
- Load your best parameter values from the parameter estimation. These parameter values should be plugged in as the
startvector in the code below. - Modify the code below such that on the line where it defines the
targetdistribution you should insert your own objectiveFunction/cost function. Please note that thetargetdistribution should be defined asexp(-f(x))wheref(x)is your objective function, asn in the example below.
While you do not have to understand the exact details of the code below, please make some effort to see if you can identify the main principles of the MCMC sampling. If you have any questions please do not hesitate to ask the supervisor.
start = parameterValues; % initial guess, use the best parameters obtained from the parameter estimation.
nsamples = 10000; % number of samples
samples = zeros(nsamples, numel(start)); % matrix to hold the samples
samples(1, :) = start; % first sample is the initial guess
% Define the target distribution based on your objective function. Note that the fileID input should be empty for now
target = @(params) exp(-objectiveFunction(params, DataStructure, modelName,[]));
burnin = 100; % number of iterations to perform before adapting proposal distribution
fixed_sigma = 0.2;
% Run the chain
for i = 2:nsamples
if i <= burnin
% For the first few iterations, use a fixed candidate distribution
candidate = normrnd(samples(i-1, :), fixed_sigma); % calculate the next candidate sample based on the previous sample.
else
% After the burn-in period, use the covariance of past samples to determine the variance of the candidate distribution
sigma = cov(samples(1:i-1, :));
candidate = mvnrnd(samples(i-1, :), sigma); % calculate the next candidate as a random perturbation of the previous sample, with sigma based the covariance of all previous samples.
end
% Compute acceptance probability
accept_prob = min(1, target(candidate)/target(samples(i-1, :)));
% Accept or reject the candidate based on the acceptance probability. This probability is based on if the objective function value of the candidate vector in relation to the objective function value of the previous sample.
if rand < accept_prob
objectiveFunction(candidate, DataStructure, modelName,fileID); %if the candidate vector is accepted call the objective function with the fileID input to save the candidate.
samples(i, :) = candidate;
else
samples(i, :) = samples(i-1, :);
end
end
If you get the ERROR: SIGMA must be a positive semi-definite matrix
NOTE! The code above dose contain some random elements and will therefore on some occasions result in the following error. If you encounter this error it is most likely due to numerical precision error and it should not persist if you run the script again. Due to numerical precision or certain characteristics of the data, the computed covariance might not be positive semi-definite.
Error using mvnrnd (line 115)
SIGMA must be a positive semi-definite matrix.
Error in simpleMCMCsampling (line 34)
step = mvnrnd(zeros(size(samples(i-1, :))), sigma);
sigma. i.e. on the line after you calculate sigma you add:
sigma = sigma + 1e-25 * eye(size(sigma)); % Add a small constant to the diagonal
Once the sampling is done you can load the saved acceptable variables with the command:
MCMCresults = load(FileName);
fileName is the string that you assigned as the file name. If you saved your parameters as described above MCMCresults will have 9 columns. The first will contain the objective function values and the following 8 will contain the respective parameter values.
Plotting parameter distributions
Now that you hopefully have some distributions of acceptable parameter values. You should plot them in a meaningful way. You are of course free to do this in any way you seam suitable. However, perhaps the most common way to illustrate a distribution of values is through the use of a histogram. Matlab's built in histogram(X) function will create a histogram for the vector X. Note that if X is a matrix the histogram function will make a single histogram for the entire matrix. This means that if you have saved the MCMC results as described above you'll want to make one histogram for each column of the file with your saves acceptable parameter values.
Please note that the implementation of the MCMC sampling algorithm that is presented above is still fairly rudimentary and does not sample the parameter space in a comprehensive fashion. Rather it is a simpler implementation for teaching the core principles of MCMC sampling for the purposes of this computer exercise. If you are interested in an implementation of a proper MCMC sampling algorithm one such example can be found here: MCMCexample. This implementation uses the PESTO-toolbox which can be found and downloaded at the provided link. You are free to implement this algorithm in this exercise but running the sampling will likely take more than an hour for a reasonable number of samples.
- The parameter distributions for a comprehensive MCMC sampling can be found in the zip file available at the link above. This example contains a complete sample file, with all acceptable parameter values. To load this file and access the parameter distributions you can simply follow the example below. Note that this file is fairly large and loading it should take a few seconds.
- Load the results and plot the parameter distributions for the comprehensive MCMC sampling.
result = load('MCMC_bp_age_230619_085348.dat');
Profile Likelihood¶
Background: Reverse Profile Likelihood
Profile Likelihood analysis is a computational approach used in model-based analysis to estimation of confidence intervals for parameters in a model, often used in the field of systems biology and other areas that employ complex mathematical models. Here we introduced what is know as a partial or some times Reverse Profile Likelihood, the full profile Likelihood analysis is introduced in more detail in the section below. The reversed profile Likelihood is as the name suggests not as comprehensive but less computationally demanding.
We can utilize an optimization algorithm to actively explore the highest and lowest permissible values for a particular model parameter without exceeding the established \(\chi^2\)-threshold. Essentially, each parameter should have some wiggle room to vary from the optimal solution before it pushes the result beyond this threshold. This range of variation for a parameter, while still contributing to an acceptable model solution, is sometimes referred to as the parameter's profile in relation to the likelihood (or cost) function.
Some parameters may have a broad range because changes can be offset by other parameters. On the other hand, for some parameters, the acceptable values range will be narrow, implying that these parameters significantly influence the model's behaviors.
Since the optimization solvers we utilized in the previous section aim to identify the parameters that minimize the given objective function, these algorithms can be repurposed to find the smallest (and largest) value for a specific parameter. If we perform this optimization for each parameter, we can establish confidence intervals for all acceptable parameter values in our model.
Figure 5: an illustration of a parameters Likelihood profile and how it relates to the maximal and minimal acceptable values for the parameter
Task 7: Perform a reversed profile likelihood analysis for the model parameters
Perform a reversed profile likelihood to analyse the parameter uncertainties. To do this you should:
- Write a new objective function that looks to minimise/maximise a specific parameter value.
- Write a script for the reversed profile likelihood analysis algorithm.
- Run the analysis.
- Use the same upper and lower bounds as for the parameter estimation above.
- Plot the parameter boundaries in a reasonable way.
- Compare parameter bounds from different methods.
- Does the parameter bounds match up with the parameter bounds you found from the MCMC sampling? Why/why not?
- How reliable are the parameter bounds?
How to construct an objective function for a reversed profile likelihood analysis
If we want to use an optimisation algorithm to find the minimum or maximum value for a certain parameter we want an objective function that reflects this purpose. To design such an objective function we need 3 addition inputs argument, compared to our previous objective function. We neen information regarding which parameter we are maximising/minimising, we need to know if we are maximising or minimising, and we need to know the \(\chi^2\)-threshold we should stay under. The following function is an example of what such an objective function might look like.
function[ v ] = objectiveFunction_reversePL(parameterValues, modelName, DataStructure, parameterIdx, polarity, threshold)
cost = objectiveFunction(parameterValues,DataStructure,modelName); %calculate cost with original objective function
v = polarity*parameterValues(parameterIdx); % return the parameter value at index parameterIdx. multiply by polarity = {-1,1} to swap between finding maximum and minimum parameter value.
if cost>threshold % check if solution is under limit
v = abs(v) + (cost-threshold).^2; % add penalty if the solution is above the limit. penalty grows the more over the limit the solution is.
end
end
cost using the original objective function. HERE you want to substitute the function objectiveFunction with your original objective function. Make sure the input arguments match those of your objective function!
The function then returns the parameter values of the parameter indicated by the parameterIdx variable. parameterIdx is a scalar value with the index of the parameter we want to maximise or minimise. The input variable polarity is either \(1\) or \(-1\) and indicates if we are maximising or minimising the parameter value. Most optimisation algorithm aim to minimize their objective function however, by multiplying the output value by \(-1\) we are effectively maximising the objective function value. Hence, if polarity = 1
we are minimising the parameter value, but if polarity = -1 we are maximising the parameter value.
Lastly, the function contains an if clause that kicks in if the cost is larger that the threshold i.e. we have an unacceptable solution. If this is the case we add a penalty term to the output value v. This penalty term will be exponentially proportional to how much above the threshold the current solution is.
Writing the algorithm for the reversed profile likelihood analysis
The main algorithm for this script should:
- Loop over all parameters in the model.
- For each parameter run two optimisations, one to maximise the parameter value, and one to minimise the parameter value.
- For this optimisation you'll want to use a global algorithm that can use your optimal parameter values as a start guess.
simulannealbndis a suitable choice. - Remember that you will want to update the anonymous objective function for each iteration as the
parameterIdx, andpolarityvariables will change. i.e. you'll likely want something like:proxyObjFun = @(x)objectiveFunction_reversePL(x ,modelName, DataStructure, parameterIdx, polarity, threshold);
While the partial or reversed profile likelihood analysis above can be used to estimate the confidence intervals of the parameter values within a reasonable computational time. We would still be interested in analysing the full likelihood profile thus we will now look at what this means and how we can implement a full Profile likelihood analysis.
A full Profile likelihood analysis
The concept behind a profile likelihood analysis involves exploring how well the model fits the data as each parameter is varied, while optimizing the other parameters. Essentially, for each value of the parameter of interest, the values of the other parameters that maximise the likelihood (minimises the cost) are found. This gives a "profile" of the likelihood function for the parameter of interest. In other words one parameter is fixated at a specific value, while all other parameters are re-optimised. Then we step the fixated parameter away from the optimal value, re-optimising all other parameters for each step.
By creating a range of acceptable values (usually based on a \(\chi^2\)-threshold), a confidence interval for each parameter can be determined. This process is then repeated for each parameter in the model.
The advantage of the profile likelihood method is that it takes into account the correlations between parameters and the shape of the likelihood surface, providing more accurate confidence intervals, especially for complex, non-linear models.
Overall, Profile Likelihood analysis is a useful technique for understanding the influence of individual parameters on a model and quantifying their uncertainties. It provides insights into which parameters are well-determined by the data, which are not, and how they interact, which are crucial for model interpretation and prediction.
Compared to the reversed or partial profile likelihood analysis described in the previous section the full profile likelihood analysis aims to map the entire likelihood profile of a parameter or model property as depicted in figure 6.
Figure 6: An illustration of the parameters Likelihood profile and how it relates to different solutions for the model fitting problem
This method enables the understanding of how parameter uncertainties propagate to model outputs or predictions, thereby providing confidence intervals for these predictions.
A Profile Likelihood analysis can also be conducted for a model property or prediction, rather than a model parameter, in this case it is called a Prediction Profile Likelihood (PPL) analysis. This method enables the understanding of how parameter uncertainties propagate to model outputs or predictions, thereby providing confidence intervals for these predictions.
Task 8: Perform a comprehensive profile likelihood analysis
To complete this task you should perform a comprehensive profile likelihood analysis for a parameter of the blood pressure model, or perform a prediction profile likelihood analysis for the model variable that describes the mean arterial pressure MAP. These two options should reflect equal task and will require you to:
- Modify your ordinal objective function such that it allows you to fixate a parameter value (or time point of the simulated MAP value) you want to analyse.
- Write a script for full (prediction) profile likelihood analysis.
- Run the analysis
- Use the same upper and lower bounds as for the parameter estimation above.
- Plot the full (prediction) likelihood profile
- This figure should resemble the left part of Figure 6 above, with the fixated parameter values/MAP values on the x-axis and the objective function value on the y-axis. (You don't need to plot the simulations to the right in Figure 6)
- Evaluate the likelihood profile of the parameter or prediction.
- How does this profile compare to you previous confidence intervals?
Modify your original objective function for profile likelihood analysis
For this tasks you'll want to modify your original objective function such that you send an index variable, and a variable containing a reference value to the objective function. The index variable, PL_index in hte example below, will point to either the parameter or the model property that is the subject of the analysis. The reference value, PL_refValue below, is the value of the fixated parameter, or model property, being stepped away from the optimum.
[cost] = objectiveFunction_PL(parameterValues,DataStructure,modelName, PL_index, PL_refValue)
For a profile likelihood analysis of a parameter value we want the objective function to keep the value of a specific parameter, indicated by PL_index, to a certain reference value PL_refValue. We can do this by adding a penalty to the objective function if the difference between the parameter value indicated by PL_index and the reference value PL_refValue is too large, as is done by the if statement below.
if and(~isempty(PL_index),~isempty(PL_refValue)) %if both PL_index and PL_refValue are not empty.
cost = cost + 1e6 .* (parameterValues(PL_index)-PL_refValue).^2; %add very large penalty if the difference between a specific parameter value and a reference value is too large.
end
For a prediction profile likelihood analysis of MAP we want the objective function to keep a certain timepoint of the MAP simulation close to our reference value PL_refValue. Similarly, to the case above we can implement this as a penalty function as shown here:
if and(~isempty(PL_index),~isempty(PL_refValue)) %if both PL_index and PL_refValue are not empty.
cost = cost + 1e6 .* (modelSimulation.variablevalues(PL_index,1)-PL_refValue).^2; %add very large penalty if the difference between a specific prediction and a reference value is too large.
end
Note that for both of these cases we multiply the difference to the reference value with a very large constant 1e6, this is to ensure that any divergence from the reference value adds a large figure to the objective function value. This constant can be tuned to the specific analysis conducted.
Write an algorithm for the (prediction) profile analysis
For the algorithm of the (prediction) profile analysis you'll want to modify your original objective function in accordance with the box above. For the algorithm itself the goal is to select a parameter of model property to create the profile for, fixating this value to a reference value, and successively stepping this reference value away from the optimal solution. For this we need to:
- Select what parameter or model property to profile, this selection is up to you!
- Determine the number of steps to take away from the optimum, in either direction (up and down).
- Calculate a step size such our profile gives us useful information once all steps are done
- Calculate the reference value for each step.
- Run an optimisation for each step.
PL_index = 5;
nSteps = 10;
stepSize =
bestFval = zeros(1,nSteps*2);
count = 1;
for step = 1:nSteps
for direction = [-1,1]
PL_refValue = optimalValue + direction*step*stepSize
proxyObjFun = @(x) objectiveFunction_PL(x ,modelName, DataStructure, PL_index, PL_refValue);
[~, bestFval(count)] = simulannealbnd(proxyObjFun,optParameterValues,lb,ub,options)
count = count + 1;
end
end
simulannealbnd is used but you are free to use any optimisation solver. Although a solver that can base its start guess on an optimal solution will probably reduce computational time.
Model uncertainty¶
From the three different approaches for estimating the model parameter uncertainty above, you should now have good estimates for the parameter uncertainties in the blood pressure model. This also means that you have an estimate for all acceptable model behaviours. By plotting these different model behaviours we can visualise the model uncertainty i.e. not only showing the best possible fit to data but rather all solutions that are acceptable with regard to the \(\chi^2\)-test.
Task 9: Evaluate model simulation uncertainty
Since you now have a good estimate for all acceptable parameter values you also have an estimate for all acceptable model behaviours. In this final task you should plot these model behaviours as model simulations.
- Run model simulations for the full range of acceptable parameter values.
- Make a figure that shows the data for SBP, DBP and MAP with the simulated model uncertainty for each data set.
Plotting a shaded area in MATLAB
For the figure you create in this task you could use 'hold on' to plot all the simulations as separate lines on top of each other. However, your plot will likely be cleaner and more readable if you instead plot the largest and lowest simulated values with a shaded area in between to illustrate that all simulations fall within the shaded area. Here is an small example for how this could be achieved in MATLAB using the fill function:
simulation_lb = min(allSimulations);
simulation_ub = max(allSimulations);
fill([time fliplr(time)], [simulation_lb fliplr(simulation_ub)],'', 'FaceColor',[0 0 1],'FaceAlpha',0.5)
allSimulations is a matrix were the rows are the different simulations and columns are the different time points of each simulation. Also note that the properties FaceColor = [0,0,1] and 'FaceAlpha = 0.5` sets the colour and transparency of the filled area. You'll want to adjust these to match the colours of your data sets.
Questions for lab 2 - Parameter estimation and model uncertainty:¶
In your report, you should present answers the following questions
Task 1 - Qualitative evaluation of initial model simulation
- What can we learn about the model and experimental data from the figure made in task 1?
Task 2 - Quantitative evaluation of model fit
- What does the "cost" tell us about the models agreement to the data?
- When calculating the "cost", why do we divide the residuals by the standard deviation or SEM?
Task 3 - Parameter Estimation
- Following a parameter optimisation, can the model explain the data?
- Does the choice of optimisation algorithm affect the solution of the best parameter values? Why/Why not?
- Does the initial parameter values affect the solution of the best parameter values? Why/Why not?
- What are the pros and cons of using a global or a local algorithm form parameter estimation?
Task 4 - Statistical hypothesis testing
- According to a \(\chi^2\) - test should the model with the optimal parameters be rejected?
- What is the null hypothesis of the \(\chi^2\) - test?
- What can you conclude about the model when you reject the \(\chi^2\) - test?
- Describe some other statistical tests that could be used to reject a model hypothesis?
- When and why would these alternatives be preferable to the \(\chi^2\) - test?
Task 6 - MCMC sampling
- What are some of the insights that can be obtained from studying the parameter distributions gathered from the MCMC sampling?
- What are the difference between identifiable and unidentifiable parameters?
- What are the pros and cons of using MCMC sampling to determine parameters distributions?
Task 7 and 8 - Profile likelihood analysis
- How does the parameter bounds you obtained from the profile likelihood analyses compare to the parameter distributions you obtained from the MCMC sampling?
- How does the parameter likelihood profile correlate to the parameter distributions?
-
What are the pros and cons of using a reversed or a full profile likelihood analysis to determine parameter/model uncertainties?
-
Which of the model parameters are identifiable?
- What conclusions can you make about this model with this information?
Task 9 - Model uncertainty
- What are some important insights that can be obtained from plotting the full model uncertainty? Compared to only plotting the best solution.