Compartments¶
In this simple example, we have two models of glucose metabolism.
Please note that these models are meant to showcase the toolbox functionality, not to correctly represent the biology.
The main model controls insulin and glucose in plasma, and provides these as inputs to the submodel. The submodel controls glucose uptake in a tissue, and provides the uptake back as outputs. In this example, we use three tissue versions of the submodel: adipose, muscle, other.
In this example, the main model is not attached to a compartment, so the tissue sub models must declare that they need inputs from outside the compartment they below to, from a model not assigned to a compartment:
########## INPUTS
adipose_uptake = :uptake @ 0
Furthermore, the main model needs input from other compartments (the submodels), so we declare which compartment the inputs should come from:
########## INPUTS
adipose_uptake = adipose:uptake @0
muscle_uptake = muscle:uptake @ 0
other_uptake = other:uptake @ 0
The full model files are given below.
Main-model
########## NAME
main_model
########## METADATA
time_unit = m
########## MACROS
########## STATES
d/dt(glu) = glu_input + dng - uptake
d/dt(ins) = ins_prod - ins_clearance
glu(0) = 5.0
ins(0) = 0
########## PARAMETERS
k_ins_prod = 0.5
k_ins_clearance = 0.5
dng = 1
ins_prod_flag=0
########## VARIABLES
ins_prod = k_ins_prod*ins_prod_flag
ins_clearance = k_ins_clearance * ins
uptake = adipose_uptake + muscle_uptake + other_uptake
########## FUNCTIONS
########## EVENTS
glu_high_event = glu>5.1, ins_prod_flag, 1
glu_low_event = glu<4.9, ins_prod_flag, 0
########## OUTPUTS
glucose_plasma = glu
insulin_plasma = ins
########## INPUTS
glu_input = glucose @ 0
adipose_uptake = adipose:uptake @0
muscle_uptake = muscle:uptake @ 0
other_uptake = other:uptake @ 0
########## FEATURES
glucose_plasma = glu
insulin_plasma = ins
Sub-model
########## NAME
sub_model
########## METADATA
time_unit = m
########## MACROS
########## STATES
d/dt(glu) = uptake - utilization
glu(0) = 5.0
########## PARAMETERS
k_uptake = 0.3
k_utilization = 1
k_M = 1
########## VARIABLES
uptake = k_uptake*glu_plasma * ins_plasma/(k_M + ins_plasma)
utilization = k_utilization * glu
########## FUNCTIONS
########## EVENTS
########## OUTPUTS
uptake = uptake
########## INPUTS
glu_plasma = :glucose_plasma
ins_plasma = :insulin_plasma
########## FEATURES
glucose = glu
uptake = uptake
# Load the model class for the sub_model
SubModel = sund.import_model("sub_model")
adipose_model = SubModel(compartment="adipose", parametervalues=[0.15, 1, 1])
muscle_model = SubModel(compartment="muscle", parametervalues=[0.8, 1, 1])
other_model = SubModel(compartment="other", parametervalues=[0.05, 1, 1])
Now, we can combine the models and run the simulation (for 100 minutes):
time_vector = np.linspace(0, 100, 1000) # 100 minutes
sim = sund.Simulation(
models = [main_model, adipose_model, muscle_model, other_model],
activities = [activity],
time_vector = time_vector,
time_unit = "m"
)
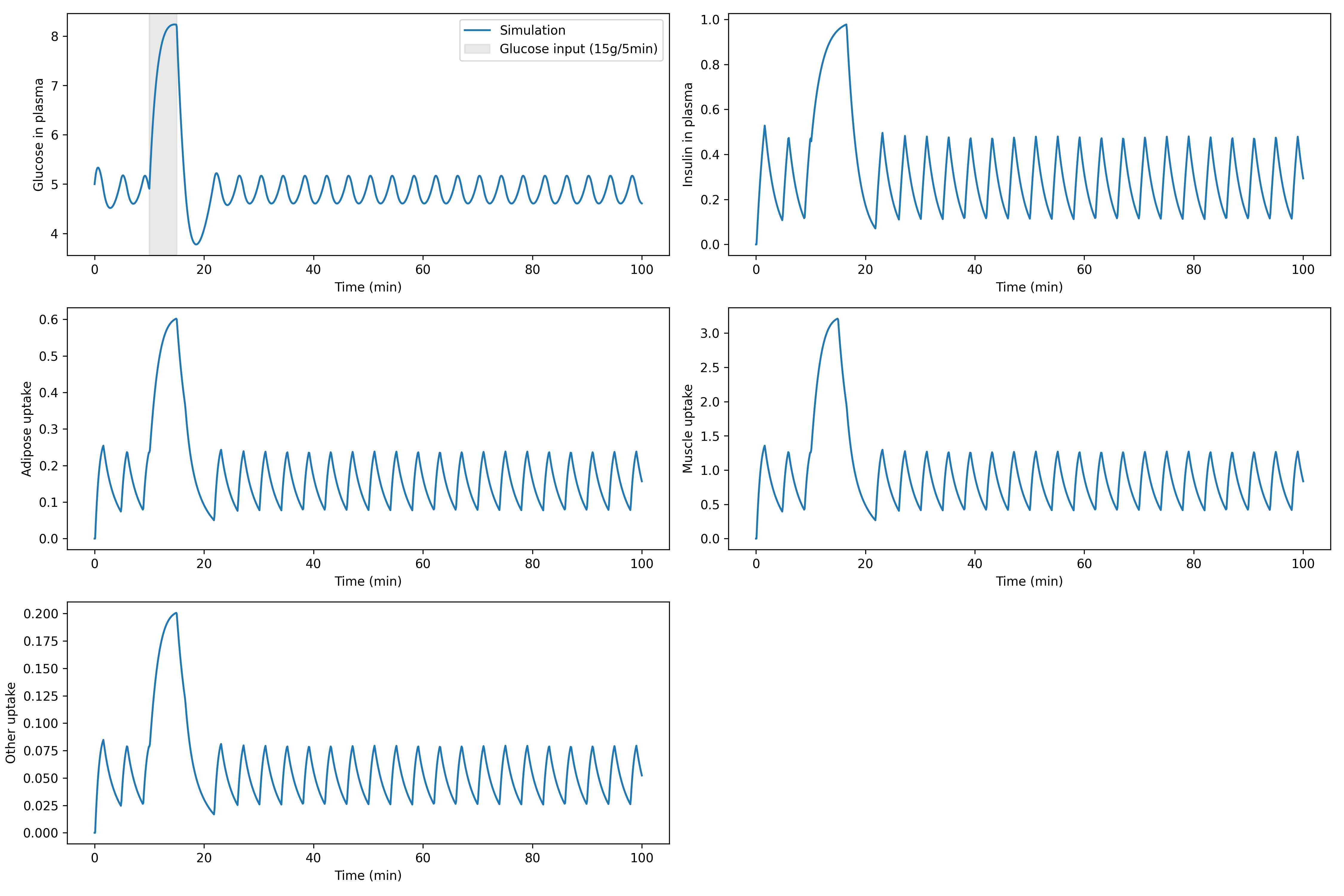
In end, we can plot the plasma glucose and insulin, and the compartment based uptakes:
Full code for the example¶
Here we provide the full code to build the compartmentalized models, simulate and plot. The code is also available here, the main model here, and the sub model here
import sund
import numpy as np
import matplotlib.pyplot as plt
sund.uninstall_model(all=True) # Uninstall all models to start fresh
# Install the models
sund.install_model("main_model.txt")
sund.install_model("sub_model.txt")
# Load the models
main_model = sund.load_model("main_model") # Load a model object directly
SubModel = sund.import_model("sub_model") # Load the model class for the sub model
adipose_model = SubModel(compartment="adipose", parametervalues=[0.15, 1, 1])
muscle_model = SubModel(compartment="muscle", parametervalues=[0.8, 1, 1])
other_model = SubModel(compartment="other", parametervalues=[0.05, 1, 1])
print(adipose_model.feature_names)
# Define the glucose input as an activity
activity = sund.Activity(time_unit="m")
activity.add_output(
name="glucose",
type="piecewise_constant",
t=[10,15],
f=[0, 15/5, 0]
) # 15 grams over 5 minutes
# Create and run the simulation
time_vector = np.linspace(0, 100, 1000) # 100 minutes
sim = sund.Simulation(
models = [main_model, adipose_model, muscle_model, other_model],
activities = [activity],
time_vector = time_vector,
time_unit = "m"
)
sim.simulate()
# Plot the results
features = sim.feature_data_as_dict()
fig, ax = plt.subplots(3, 2, figsize=(15, 10))
ax[0, 0].plot(sim.time_vector, features["glucose_plasma"], label = "Simulation")
ax[0, 0].set_ylabel("Glucose in plasma")
ax[0, 0].set_xlabel("Time (min)")
ax[0, 0].axvspan(10, 15, color='lightgrey', alpha=0.5, label="Glucose input (15g/5min)")
ax[0, 0].legend()
ax[0, 1].plot(sim.time_vector, features["insulin_plasma"])
ax[0, 1].set_ylabel("Insulin in plasma")
ax[0, 1].set_xlabel("Time (min)")
ax[1, 0].plot(sim.time_vector, features["adipose:uptake"])
ax[1, 0].set_ylabel("Adipose uptake")
ax[1, 0].set_xlabel("Time (min)")
ax[1, 1].plot(sim.time_vector, features["muscle:uptake"])
ax[1, 1].set_ylabel("Muscle uptake")
ax[1, 1].set_xlabel("Time (min)")
ax[2, 0].plot(sim.time_vector, features["other:uptake"])
ax[2, 0].set_ylabel("Other uptake")
ax[2, 0].set_xlabel("Time (min)")
fig.delaxes(ax[2, 1]) # remove unused last subplot
plt.tight_layout()
plt.savefig("compartment_example.png", dpi=300)
plt.show()