Large-scale modelling
In addition to the wellstudied small-scale mechanistic models we typically work with, we have also done some modelling on a larger scale. Ideally, all processes could be studied in the same level of detail that we typically do on the small scale, however this is simply not feasible when it comes to handling thousands of features (proteins, phosphorylations, flows, etc). To that end, we have both developed novel methods for handling large-scale data while still maintaining the mechanistic characteristics, and used available methods to model the available large-scale data.
Developing methods for modelling time-series data on a large scale.
As previously mentioned, we have worked with developing methods for analyzing large-scale time-series data. In our first project, we collaborated with Mika Gustafsson, and developed a method for using a set of core transcription factors as the input to the rest of the genome.
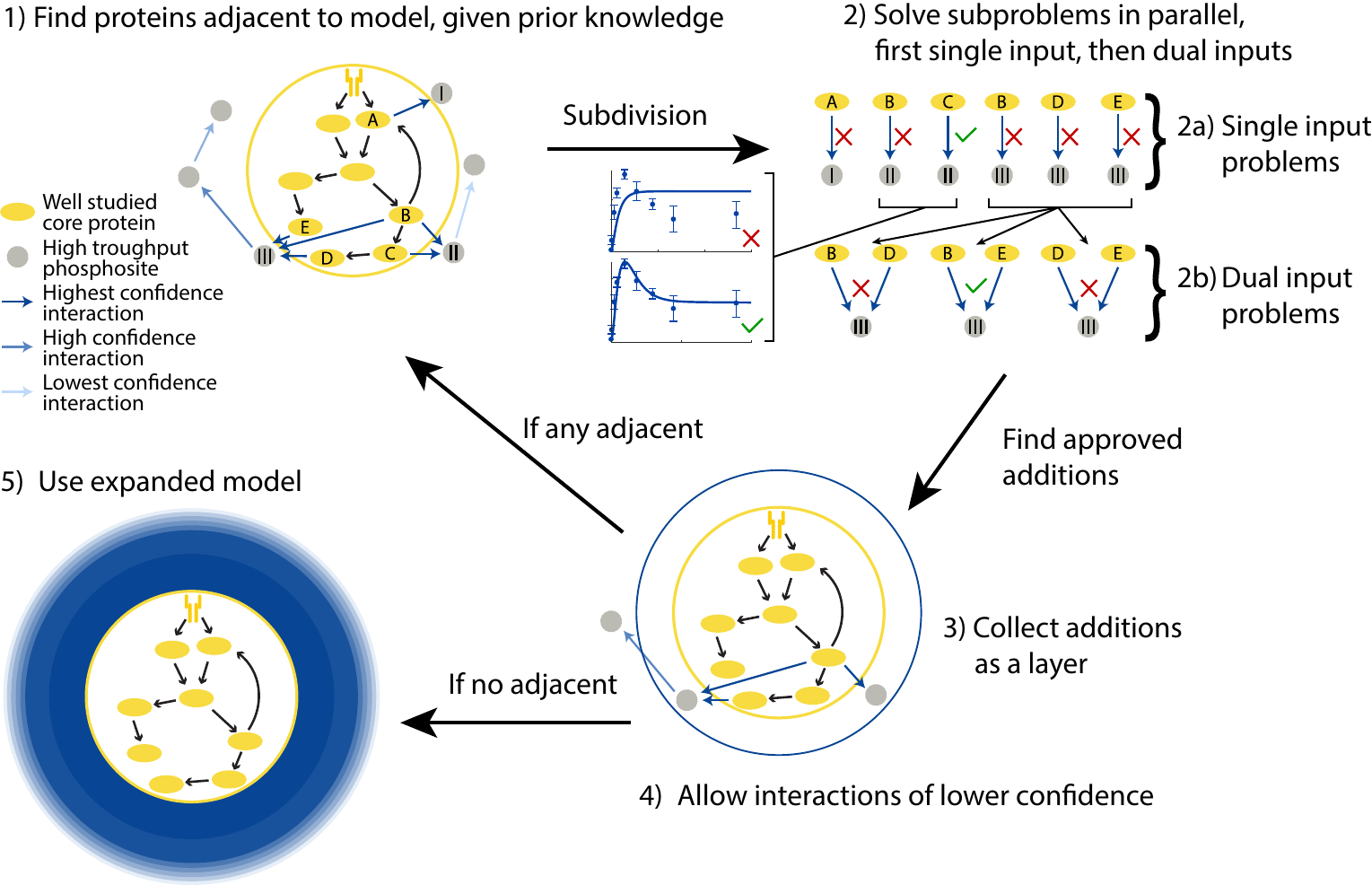
This method was later extended, when we developed a method for automatically expanding a small-scale insulin signalling model to cover a majority of the (available, measured) phosphoproteome by iteratively expanding the model (illustrated in the figure above, reused from this preprint).
- Magnusson R, Mariotti GP, Köpsén M, Lövfors W, Gawel DR, Jörnsten R, et al. LASSIM—A network inference toolbox for genome-wide mechanistic modeling. PLOS Computational Biology. 2017 Jun 22;13(6):e1005608.
- Lövfors W, Jönsson C, Olofsson CS, Nyman E, Cedersund G. A comprehensive mechanistic model of adipocyte signaling with layers of confidence. bioRxiv. 2022. p. 2022.03.11.483974.
Cancer modelling
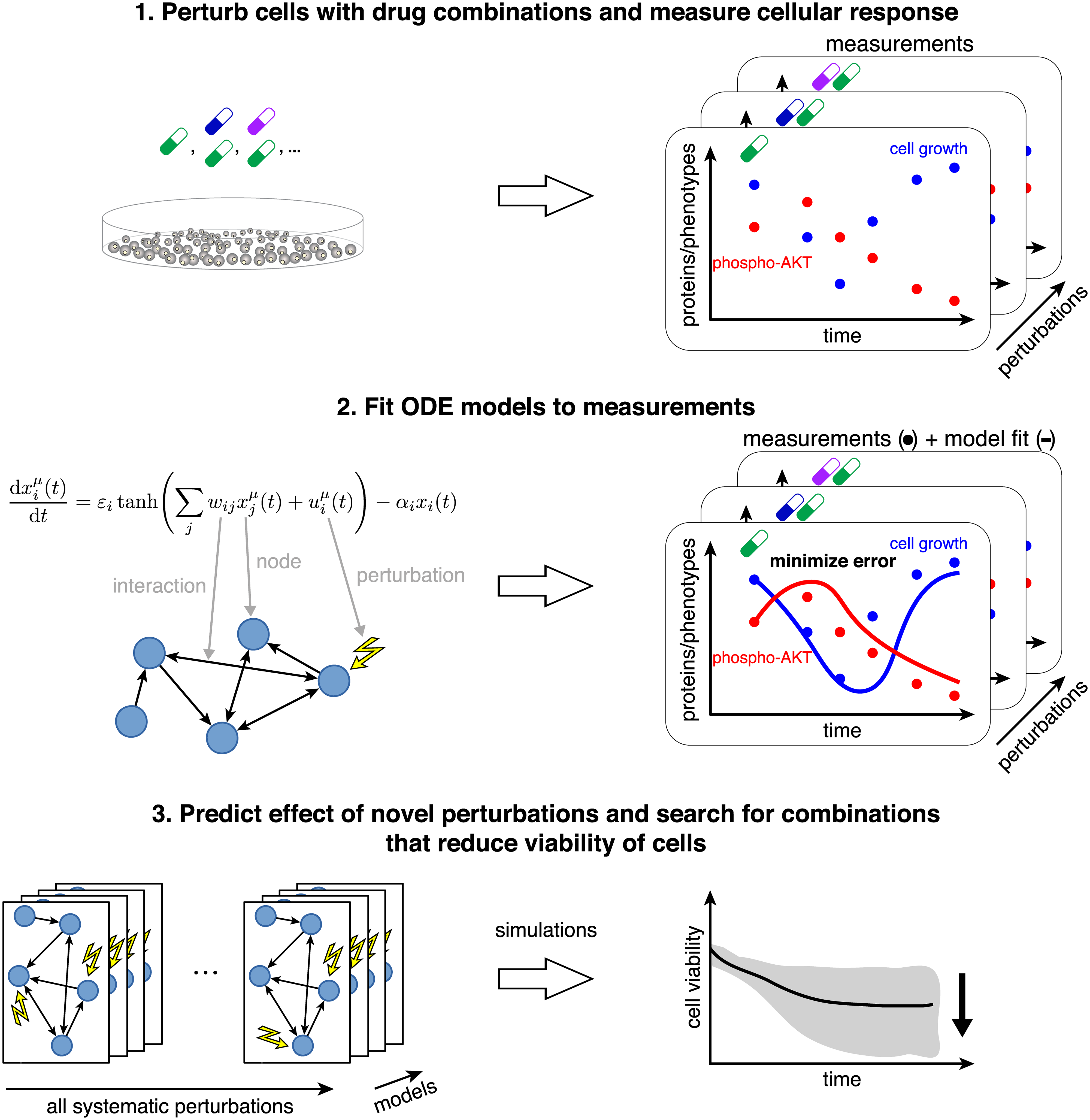
In addition to the development of novel methods of generating large-scale models, we have also used available methods to model the impact of cancer drugs. In this project, rich data containing multiple different perturbations with drugs and combinations of drugs was used. While such data are very informative, it can also be problematic to analyze without using modelling. We therefore used time-resolved mathematical modelling to couple molecular events to cellular phenotypes. The method is illustrated in the figure above (reused from Nyman et. al. 2020).
